Gplots Install
- Matplotlib Plot Function
- Install Ggplot2 R Studio
- Plot3d R Package
- Plots Install
- Gplots Install Ubuntu
- Ggplot2 Package R
gplots: Various R Programming Tools for Plotting Data
Matplotlib Plot Function
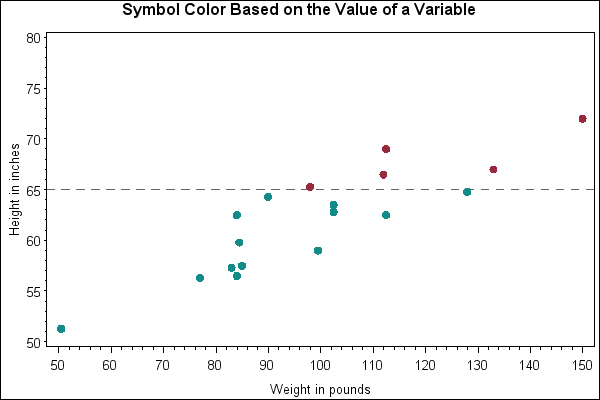

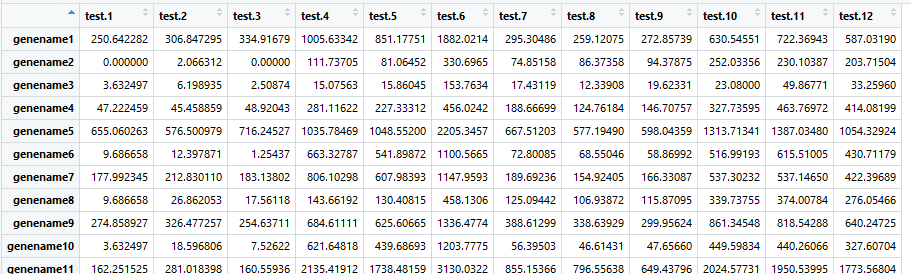
Install.packages(' gplots ') Citation of original package: Gregory R. Warnes, Ben Bolker, Lodewijk Bonebakker, Robert Gentleman, Wolfgang Huber Andy Liaw, Thomas Lumley, Martin Maechler, Arni Magnusson, Steffen Moeller, Marc Schwartz and Bill Venables (2015). Gplots: Various R Programming Tools for Plotting Data. 'gplots' packages does not work. I had installed de 'gplots' package and then I loaded the library but the package did not work. I recieved a message like this:. Install the needed R packages. Set some options and make sure the packages edgeR, gplots, RColorBrewer, topGO, KEGGREST, Rgraphviz and org.Mm.eg.db are installed (if not install it), and then load. In the R console run the following commands. If(!require(gplots)) install.packages('gplots') if(!require(LDheatmap)) install.packages('LDheatmap') if(!require(genetics)) install.packages('genetics') if(!require.
Various R programming tools for plotting data, including: - calculating and plotting locally smoothed summary function as ('bandplot', 'wapply'), - enhanced versions of standard plots ('barplot2', 'boxplot2', 'heatmap.2', 'smartlegend'), - manipulating colors ('col2hex', 'colorpanel', 'redgreen', 'greenred', 'bluered', 'redblue', 'rich.colors'), - calculating and plotting two-dimensional data summaries ('ci2d', 'hist2d'), - enhanced regression diagnostic plots ('lmplot2', 'residplot'), - formula-enabled interface to 'stats::lowess' function ('lowess'), - displaying textual data in plots ('textplot', 'sinkplot'), - plotting a matrix where each cell contains a dot whose size reflects the relative magnitude of the elements ('balloonplot'), - plotting 'Venn' diagrams ('venn'), - displaying Open-Office style plots ('ooplot'), - plotting multiple data on same region, with separate axes ('overplot'), - plotting means and confidence intervals ('plotCI', 'plotmeans'), - spacing points in an x-y plot so they don't overlap ('space').

Install Ggplot2 R Studio
| Version: | 3.1.1 |
| Depends: | R (≥ 3.0) |
| Imports: | gtools, stats, caTools, KernSmooth |
| Suggests: | grid, MASS, knitr |
| Published: | 2020-11-28 |
| Author: | Gregory R. Warnes [aut], Ben Bolker [aut], Lodewijk Bonebakker [aut], Robert Gentleman [aut], Wolfgang Huber [aut], Andy Liaw [aut], Thomas Lumley [aut], Martin Maechler [aut], Arni Magnusson [aut], Steffen Moeller [aut], Marc Schwartz [aut], Bill Venables [aut], Tal Galili [ctb, cre] |
| Maintainer: | Tal Galili <tal.galili at gmail.com> |
| BugReports: | https://github.com/talgalili/gplots/issues |
| License: | GPL-2 |
| URL: | https://github.com/talgalili/gplots |
| NeedsCompilation: | no |
| Materials: | |
| In views: | Graphics |
| CRAN checks: | gplots results |
Downloads:
Plot3d R Package
| Reference manual: | gplots.pdf |
| Vignettes: | Venn Diagrams with gplots |
| Package source: | gplots_3.1.1.tar.gz |
| Windows binaries: | r-devel: gplots_3.1.1.zip, r-release: gplots_3.1.1.zip, r-oldrel: gplots_3.1.1.zip |
| macOS binaries: | r-release: gplots_3.1.1.tgz, r-oldrel: gplots_3.1.1.tgz |
| Old sources: | gplots archive |
Reverse dependencies:
Plots Install
| Reverse depends: | bayess, cellVolumeDist, COHCAP, corkscrew, DandEFA, DensParcorr, difconet, EBSeq, GSCA, HD2013SGI, highSCREEN, Hiiragi2013, iCheck, iCluster, made4, massiR, MBttest, MDplot, MetabolAnalyze, Meth27QC, MethTargetedNGS, mgsa, mlma, MLP, mma, MOQA, muma, NBBttest, NestLink, PRISMA, RCA, ReorderCluster, RnBeads, SCINA, swamp, SwimR, switchBox, tRanslatome |
| Reverse imports: | a4Base, ABAEnrichment, affycoretools, ArchaeoPhases, artMS, ASSIGN, asymmetry, BAMMtools, BatchQC, bingat, Biopeak, BNPMIXcluster, boundingbox, BUScorrect, cancerTiming, cbaf, CellScore, cellTree, CHETAH, ChIPseeker, chromswitch, CINdex, CluMSID, CNVPanelizer, CoFRA, CoGAPS, cogena, colordistance, compcodeR, compEpiTools, consensus, contiBAIT, corregp, corto, covid19.analytics, crmReg, crqa, DAPAR, debrowser, deco, DepecheR, DeSousa2013, dfConn, DiffBind, DiffNet, DmelSGI, DNMF, DPBBM, eegc, EGAD, EGSEA, ENmix, erccdashboard, expands, fDMA, flagme, Fletcher2013a, FLLat, flowCyBar, flowDensity, GDCRNATools, GeneOverlap, GOexpress, graphsim, haploReconstruct, HIREewas, HMP, HTqPCR, ideal, ImmuneSpaceR, indirect, infercnv, IntClust, ionr, isomiRs, kissDE, KSEAapp, les, liayson, LineagePulse, lmSupport, M3Drop, MAIT, matie, metagene, metagenomeSeq, MetaIntegrator, MetaNeighbor, metaseqR, metaseqR2, methylPipe, MetProc, miRLAB, mitch, MLZ, mmabig, mogsa, momr, MoonlightR, msmsEDA, MSstats, NCA, nearBynding, netboxr, oneSENSE, optCluster, PAA, Patterns, phenoTest, piano, plotMCMC, poliscidata, polyPK, pwOmics, qat, quantable, r.jive, r4ss, RAM, rcellminer, RcmdrPlugin.BiclustGUI, rdi, ReactomeGSA, Repitools, rexposome, rgsepd, RITAN, RNAdecay, RNAinteract, RobMixReg, robustSingleCell, ROCR, RPPanalyzer, SAMtool, scone, SCOPE, seriation, serieslcb, sesem, shapeR, sigQC, SingleCellSignalR, sourceR, spatialHeatmap, SpidermiR, sRACIPE, STATegRa, STRINGdb, TFARM, tigre, TIMP, tmod, TopKLists, TransView, Trendy, TSCAN, uSORT, variancePartition, vulcan, washeR, wilson, yarn |
| Reverse suggests: | AgiMicroRna, BioQC, CancerInSilico, ChemmineR, ChIPpeakAnno, clusternomics, cola, compendiumdb, condvis, CoRegNet, CSFA, CTD, DeepBlueR, dendextend, dendsort, DGCA, enveomics.R, FRESA.CAD, gmodels, heatmaply, heplots, HistData, httk, interactiveDisplay, isobar, limma, Linnorm, matR, MCbiclust, metamicrobiomeR, MKmisc, MSnbase, multiClust, OmicsPLS, Onassis, PAFway, psichomics, PtH2O2lipids, QFeatures, rattle, RforProteomics, rich, RMassBank, sensitivity, sim1000G, simba, Single.mTEC.Transcriptomes, STAN, TargetScore, TargetScoreData, timeOmics, TOAST, varrank |
Gplots Install Ubuntu
Linking:
Ggplot2 Package R
Please use the canonical formhttps://CRAN.R-project.org/package=gplotsto link to this page.